The genetic enhancement of any crop, including finger millet, requires a certain degree of genetic variation for effective utilization in crop improvement programs. However, in Ethiopia, there is limited information on the extent and pattern of genetic variability of finger millet collections under diversified agro-climatic conditions. This makes it difficult for a population to adapt to changing environmental conditions. As a result, the population may be more vulnerable to extinction, exposure to new diseases, low productivity, and selection acting on any genes that may provide disease resistance. Therefore, knowledge of genetic variability is crucial for breeders in order to develop new cultivars with desired traits that are beneficial for both farmers and breeders. The present investigation was carried out to estimating the genetic variability, heritability, and genetic advance in sixty four finger millet accessions for yield and yield-related traits at Mechara agricultural research center. The experimental design was laid out in 8 x 8 simple lattice design. The analysis of variance for mean sum of squares due to genotypes revealed highly significant differences for all the 17 quantitative characters. The genotypes showed the highest mean performance for grain yield ranged from 1.38 ton per hectare for ACC#208448 to 4.35 ton per hectare for ACC#230255. Whereas, genotypic and phenotypic coefficients of variation were found high for biomass yield, harvest index, and grain yield. Broad-sense heritability ranged from 50.12% for the number of fingers per ear to 93.18% for days to heading. High heritability coupled with high genetic advance as percent of mean were observed for leaf number, finger length, ear weight, thousand grain weight, biomass yield, and harvest index. In general, the results demonstrated that the finger millet accessions exhibited a high degree of genetic variability for the traits studied, which can be helpful for genetic enhancement.
| Published in | International Journal of Biomedical Science and Engineering (Volume 12, Issue 1) |
| DOI | 10.11648/j.ijbse.20241201.12 |
| Page(s) | 10-18 |
| Creative Commons |
This is an Open Access article, distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution and reproduction in any medium or format, provided the original work is properly cited. |
| Copyright |
Copyright © The Author(s), 2024. Published by Science Publishing Group |
Heritability, Finger Millet, Genetic Advance, Genetic Variability
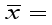 Grand mean.
Grand mean. S/N | Accession number | Collection region | Alti. | Long | Lati. |
|---|---|---|---|---|---|
1 | ACC#244798 | SNNPR | 2169 | 37.9 | 7.3 |
2 | ACC#243644 | Amhara | 1815 | 36.6 | 11 |
3 | ACC#243638 | Amhara | 1870 | 37.3 | 12 |
4 | Ikhulule | Released | |||
5 | ACC#245088 | Oromia | 2060 | 37.2 | 9.8 |
6 | ACC#243640 | Amhara | 1890 | 36.8 | 11 |
7 | ACC#243637 | Amhara | 1870 | 37.3 | 12 |
8 | ACC#245092 | Oromia | 1954 | 36.4 | 8.5 |
9 | ACC#237969 | Oromia | 1930 | 37.6 | 9.8 |
10 | ACC#237583 | Oromia | 1990 | 38.6 | 7.2 |
11 | ACC#238303 | Tigray | 2020 | 39.6 | 13 |
12 | ACC#238337 | Tigray | 1920 | 38.1 | 14 |
13 | ACC#238320 | Tigray | 2020 | 38.1 | 14 |
14 | ACC#238297 | Tigray | 2000 | 38.1 | 14 |
15 | ACC#238333 | Tigray | 1110 | 38.2 | 14 |
16 | ACC#238306 | Tigray | 2000 | 38.1 | 14 |
17 | ACC#215908 | Amhara | 2250 | 36.9 | 11 |
18 | ACC#215976 | Amhara | 1860 | 37.3 | 12 |
19 | Meba | Released | |||
20 | ACC#215968 | Amhara | 2500 | 37.5 | 13 |
21 | ACC#240506 | Amhara | 1880 | 37.7 | 11 |
22 | ACC#216033 | Oromia | 1930 | 35.7 | 9.3 |
23 | ACC#215994 | Amhara | 2050 | 37.7 | 12 |
24 | ACC#215889 | Amhara | 2100 | 37.1 | 11 |
25 | Kumsa | Released | |||
26 | ACC#235141 | Amhara | 1870 | 37.4 | 12 |
27 | ACC#234202 | Tigray | 2050 | 38.5 | 14 |
28 | ACC#237468 | Tigray | 1940 | 38 | 14 |
29 | ACC#234198 | Tigray | 1900 | 38.3 | 14 |
30 | ACC#237463 | Tigray | 2080 | 38.8 | 14 |
31 | ACC#237452 | Tigray | 1430 | 38.8 | 14 |
32 | ACC#234208 | Tigray | 1950 | 37.7 | 14 |
33 | ACC#216055 | Oromia | 1600 | 35.3 | 9 |
34 | ACC#216035 | Oromia | 1900 | 35.7 | 9.3 |
35 | ACC#219818 | Tigray | 2260 | 38.9 | 14 |
36 | ACC#216048 | Oromia | 1640 | 35.2 | 9.7 |
37 | ACC#219807 | Tigray | 1880 | 38.7 | 14.2 |
38 | ACC#216049 | Oromia | 1600 | 35.1 | 9.8 |
39 | ACC#216052 | Oromia | 1660 | 35.6 | 9.1 |
40 | ACC#216037 | Oromia | 1950 | 35.6 | 9.4 |
41 | ACC#228304 | Amhara | NA | 37.7 | 13 |
42 | ACC#234187 | Tigray | 1850 | 38.2 | 14.1 |
43 | ACC#229722 | B- Gumuz | 1750 | 36.7 | 11.2 |
44 | ACC#219824 | Tigray | 1920 | 38.3 | 14.2 |
45 | ACC#234175 | Tigray | NA | 38.1 | 14 |
46 | ACC#229726 | B-Gumuz | 1600 | 36.2 | 10.7 |
47 | ACC#230255 | B-Gumuz | NA | 36.7 | 11.2 |
48 | ACC#228902 | Oromia | NA | 36.2 | 8.6 |
49 | ACC#215869 | Amhara | 2260 | 37.4 | 11.4 |
50 | ACC#208724 | Oromia | 1640 | 37.6 | 9.8 |
51 | ACC#208448 | Amhara | 1880 | 36.4 | 11.1 |
52 | ACC#212694 | Amhara | 2380 | 38 | 11.8 |
53 | ACC#208726 | Oromia | 1880 | 36.8 | 8.5 |
54 | ACC#215883 | Amhara | 2400 | 37.7 | 11.1 |
55 | ACC#208446 | Amhara | 1920 | 37.4 | 12.4 |
56 | ACC#215873 | Amhara | 2330 | 37.4 | 11.4 |
57 | ACC#240506 | SNNPR | NA | 35.8 | 7.3 |
58 | ACC#242131 | Amhara | 2350 | 37.4 | 12.5 |
59 | ACC#242105 | Amhara | 1860 | 37.6 | 11.2 |
60 | ACC#243617 | Amhara | 1780 | 39.8 | 11 |
61 | ACC#242628 | Tigray | 1740 | 39.6 | 14.1 |
62 | ACC#241769 | SNNPR | 1500 | 37.5 | 5.5 |
63 | ACC#242618 | Tigray | 1950 | 39.6 | 14.6 |
64 | ACC#242620 | Tigray | 1770 | 38.4 | 14.8 |
Traits | Rep (1) | Genotype (63) | Blocks with in rep (Adj) (14) | Error | Total (127) | RE to RCBD | R2 | CV% | |
|---|---|---|---|---|---|---|---|---|---|
Intra (49) | RCBD (63) | ||||||||
DH | 27.2* | 166.57** | 7.52 | 5.41 | 5.88 | 85.76 | 102.28 | 0.98 | 2.52 |
DM | 63.28ns | 131.94** | 18.83 | 16.47 | 17.00 | 74.38 | 100.39 | 0.91 | 2.79 |
NL | 5.24ns | 12.11** | 2.11 | 1.43 | 1.58 | 6.83 | 103.20 | 0.92 | 9.62 |
NT | 3.063* | 2.41** | 0.35 | 0.69 | 0.61 | 1.52 | 89.11 | 0.83 | 9.89 |
NPT | 6.71** | 0.44** | 0.15 | 0.13 | 0.13 | 0.34 | 100.49 | 0.85 | 6.53 |
PH | 1320** | 319.21** | 129.63 | 81.67 | 92.33 | 214.55 | 104.46 | 0.85 | 12.10 |
NFPE | 0.79ns | 3.90** | 1.42 | 1.26 | 1.30 | 2.58 | 100.34 | 0.81 | 16.00 |
FL | 3.45ns | 3.72** | 1.05 | 0.82 | 0.87 | 2.30 | 101.35 | 0.86 | 9.36 |
Fwd | 0.269* | 0.25** | 0.07 | 0.06 | 0.07 | 0.16 | 100.05 | 0.84 | 11.6 |
NEPP | 1.14ns | 2.05** | 0.63 | 0.60 | 0.62 | 1.33 | 99.86 | 0.82 | 17.1 |
EL | 7.01** | 4.11** | 1.03 | 0.89 | 0.92 | 2.55 | 100.49 | 0.87 | 9.04 |
Ewd | 0.08ns | 1.09** | 0.30 | 0.25 | 0.26 | 0.67 | 100.57 | 0.85 | 9.46 |
EW | 0.02ns | 3.47** | 0.18 | 0.23 | 0.22 | 1.83 | 94.86 | 0.95 | 5.74 |
TSW | 0.03ns | 0.65** | 0.07 | 0.12 | 0.11 | 0.38 | 90.69 | 0.87 | 11.6 |
BMY | 0.01ns | 22.00** | 4.72 | 4.07 | 4.21 | 13.90 | 100.48 | 0.88 | 17.8 |
HI | 72.86* | 117.75** | 17.25 | 10.97 | 12.36 | 37.16 | 104.28 | 0.89 | 16.56 |
GY | 1.82* | 1.23** | 0.58 | 0.34 | 0.39 | 0.81 | 106.02 | 0.84 | 20.14 |
Traits | Min | Max | SE | Mean | MSg | σ2e | σ2g | σ2p | GCV% | PCV% | H2% | GA | GAM |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
DH | 73.50 | 107.00 | 1.71 | 92.30 | 166.57 | 5.88 | 80.34 | 86.22 | 9.71 | 10.06 | 93.18 | 17.85 | 19.34 |
DM | 129.00 | 164.50 | 2.92 | 145.60 | 131.94 | 17.00 | 57.47 | 74.47 | 5.21 | 5.93 | 77.18 | 13.74 | 9.44 |
LN | 7.25 | 18.25 | 0.89 | 12.43 | 12.11 | 1.58 | 5.26 | 6.85 | 18.46 | 21.05 | 76.89 | 4.15 | 33.39 |
NT | 6.90 | 10.80 | 0.55 | 9.89 | 2.41 | 0.61 | 0.90 | 1.51 | 9.57 | 12.43 | 59.31 | 1.50 | 15.21 |
NPT | 4.60 | 6.95 | 0.26 | 5.52 | 0.44 | 0.13 | 0.15 | 0.29 | 7.08 | 9.71 | 53.18 | 0.59 | 10.66 |
PH | 46.30 | 102.30 | 6.79 | 74.68 | 319.21 | 92.33 | 113.44 | 205.77 | 14.26 | 19.21 | 55.13 | 16.31 | 21.85 |
NFPE | 3.40 | 10.30 | 0.80 | 7.02 | 3.90 | 1.30 | 1.30 | 2.60 | 16.25 | 22.95 | 50.12 | 1.67 | 23.73 |
FL | 5.30 | 11.80 | 0.66 | 9.65 | 3.72 | 0.87 | 1.43 | 2.29 | 12.37 | 15.69 | 62.17 | 1.94 | 20.13 |
Fwd | 1.69 | 3.43 | 0.18 | 2.20 | 0.25 | 0.07 | 0.09 | 0.16 | 13.84 | 18.07 | 58.67 | 0.48 | 21.87 |
NEPP | 1.90 | 6.90 | 0.56 | 4.54 | 2.05 | 0.62 | 0.72 | 1.33 | 18.64 | 25.43 | 53.72 | 1.28 | 28.18 |
EL | 5.95 | 12.45 | 0.68 | 10.41 | 4.11 | 0.92 | 1.60 | 2.51 | 12.14 | 15.23 | 63.48 | 2.08 | 19.95 |
Ewd | 3.55 | 6.35 | 0.36 | 5.33 | 1.09 | 0.26 | 0.41 | 0.68 | 12.08 | 15.46 | 61.07 | 1.04 | 19.48 |
EW | 5.90 | 11.80 | 0.33 | 8.34 | 3.47 | 0.22 | 1.63 | 1.85 | 15.30 | 16.29 | 88.21 | 2.47 | 29.64 |
TSW | 1.53 | 3.89 | 0.24 | 3.04 | 0.65 | 0.11 | 0.27 | 0.38 | 17.07 | 20.33 | 70.50 | 0.90 | 29.57 |
BMY | 4.62 | 18.10 | 1.45 | 11.30 | 22.00 | 4.21 | 8.90 | 13.11 | 26.39 | 32.04 | 67.87 | 5.07 | 44.86 |
HI | 14.86 | 44.44 | 3.63 | 26.92 | 117.75 | 26.41 | 45.67 | 72.08 | 25.10 | 31.54 | 63.36 | 11.10 | 41.22 |
GY | 1.430 | 4.36 | 0.44 | 2.88 | 1.23 | 0.39 | 0.42 | 0.81 | 22.50 | 31.23 | 51.90 | 0.96 | 33.44 |
| [1] | Abhilash PV, Sirisha Rani B, Suresh BG and, Jalandhar Ram. 2020. Correlation and path analysis studies in finger millet for yield and yield contributing traits Int J Chem Stud, 8(6): 1696–1701. |
| [2] | Abunu Marefia, Alemu Abate, and Muluken Bantayehu. 2022. Genetic Gain in Yield Potential and Related Traits of Finger Millet in Ethiopia. East African Journal of Sciences, 16(2): 155–170. |
| [3] | Allard R. W,. 1960. Principles of plant breeding John Wiley and Sons, Inc New York. 48-49. |
| [4] | Andualem Wolie, Tadesse Dessalegn, and Ketema Belete. 2013. Heritability, variance components, and genetic advance of some yield and yield-related traits in finger millet Genotypes. African Journal of Biotechnology, 12(36): 5529–5534. |
| [5] | Anteneh Damot Mekbib Firew, Tadesse Taye, and Dessalegn Yigzaw. 2019. Genetic Diversity among Lowland Finger Millet Accessions. Ethiopia. J. Agric. Sc., 29(2): 93-108. |
| [6] | Anusha Udamala, B. Vijayalakshmi, N. Anuradha, K. Patro, and Sekhar. 2020. Studies on Genetic Variability for Yield and Quality Traits in Finger Millet. Int. J. Curr. Microbiol. App. Sci., 9(09): 641-649. |
| [7] | Brhane, H., Haile Selassie, T., and Tesfaye, K. (2017). Genetic diversity and population structure of Ethiopian finger millet genotypes using inter simple sequence repeat (ISSR) markers. Afr. J. Biotechnology. 16, 1203–1209. |
| [8] | Burton G. W. and Devane E. M. 1953, "Estimating heritability in tall fescue from replicated clonal material," Agronomy Journal, 45: 478–481. |
| [9] | Dagnachew Lule, Kassahun Tesfaye, Masresha Fetene, and Santie De Villiers. 2012, Inheritance and Association of Quantitative Traits in Finger Millet. International Journal of Genetics, 2(2): 12–21. |
| [10] | Dagnachew Lule. 2015. Assessment of Genetic Diversity, Genotype by Environment Interaction, Blast Disease Resistance, and Marker Development for Finger Millet Germplasm from Ethiopia and Introduced: 1-195. |
| [11] | Damtie Yaregal, Girma Firezer, Terfessa Alemu, and Demelash Habtamu. 2019. Genetic Diversity and Heritability Estimates among Ethiopian Finger Millet Genotypes for Yield and their Contributing Traits at Assosa, Western Ethiopia. Asian Journal of Plant Science and Research, 9(2): 6–15. |
| [12] | Debara Mekonen and Bekele Merkinel. 2021. Pre-scaling up of an improved finger millet variety at Weyira district, under Halaba zone, in Ethiopia. J Agaric Sc Food Technol, 7(3): 297–301. |
| [13] | Falconer DS. 1996. Introduction to quantitative genetics. Pearson Education India. |
| [14] | Gaertn, L. 2018. Studies on genetic variability for yield and yield-attributing traits in the finger. |
| [15] | Gohel DS and Chaudhari SB. 2018. Study of correlation and path analysis of finger millet genotypes. Journal of Pharmacognosy and Phytochemistry, 7(6): 128-1288. |
| [16] | Hilu KW. 1994. Validation of the combination Eleusine coracana subspecies africana (Kennedy O’Byrne: 410–41. |
| [17] | Hilu, K. W.; de Wet, J. M. J.; Harlan, J. R. 1979. "Archaeology botanical Studies of Eleusine coracana ssp. coracana" American Journal of Botany. 66(3): 330–333. |
| [18] | Hiremath SC, and Salimath SS., 1992. The A genome donor of Theoretical and Applied Genetics. 84: 747-754. |
| [19] |
IBPGR. 1985. Descriptors for finger millet Rome, Italy: International Board for Plant Genetic Resources. 20:
http://www2.bioversityinternational.org/publications/Web_version/417/ |
| [20] | Johnson, H., Robinson, H. F., and Comstock, R. E. 1955. Genotypic and phenotypic correlations in soybean and their implication in selection. Agronomy Journal, 47: 477–483. |
| [21] | Kebede D., Dagnachew Lule, Megersa D., Chemeda B., M. Girma M., and Geleta G. 2019. Genotype by Environment Interaction and Grain Yield Stability of Ethiopian Black-Seeded Finger Millet Genotypes. African Crop Science Journal, 27(2): 281–294. |
| [22] | Keerthana K., Chitra S., Subramanian A., and Elangovan M. 2019. Character association and path coefficient analysis in finger millet genotypes under sodic conditions. The Pharma Innovation Journal, 8(6): 556–559. |
| [23] | Khorgade, P., Narkhede, W. and Raut, S. 1985. Genetic variability in chickpea. International Chickpea Newsletter, No. 5. 3-4. |
| [24] | Madhavilatha, L., Sudhakar, P., Latha, P., Priya, M. S., and Hemanth, M. 2021. Studies on genetic variability, correlation and path analysis for quantitative traits in finger millet. 10(6), 709–712. |
| [25] | Mahanthesha, M., M. Sujatha, Ashok Kumar, and S. R. Pandravada. 2017. Studies on Variability, Heritability, and Genetic Advance for Quantitative Characters in Finger Millet Germplasm. Int. J. Curr. Microbiol. Ap. Sci., 6(6): 970–974. |
| [26] | Odeny DA. The potential of finger millet (Eleusine coracana L.) as a multi-nutrient crop for inclusion in food production systems in Africa. African Journal of Food, Agriculture, Nutrition and Development. 2013; 13(5). |
| [27] | Rohit, A. S. Jeena, Wanna Soe, Divya Chaudhary, and Ankit Kumar. 2021. Genetic Parameters Assessment of Yield-Attributing Traits in Finger Millet Germplasm Collected from September 14–17. |
| [28] | Sharma S, Vasistha NK, Meena VS. Ragi: A nutritious cereal for future. Agriculture and Biology Journal of North America. 2011; 2(9): 1302-1307. |
| [29] | Shashank A Tidke. 2020. Neutraceuticals Potential of Finger millet: Review. Journal of Pathology Research Reviews and Reports. SRC/JPR-113. 112. |
| [30] | Singh RK and Choudhury BD. 1985. "Biometrical method in quantitative genetic analysis." Kalyani Publishers, Ludhiana and New Delhi, 54–57. |
| [31] | Singh, B. D. 2005. Plant Breeding: Principles and Methods. Kalyani publishers, 7thed. New Delhi, India. |
| [32] | Sivasubramanian S. and Madhavamenon P. 1973. “Combining ability in rice,” Madras Agricultural Journal, vol. 60. 419–421. |
| [33] | Tafere Mulualem. 2022. Studies on the genetic variability, heritability, and genetic advance of finger millet. Journal of Current Opinion in Crop Science, 3(2): 55–61. |
| [34] | Udamala, A., Vijayalakshmi, B., Anuradha, N., Patro, T. S. S. K., and Sekhar, V. 2020. Studies on Genetic Variability for Yield and Quality Traits in Finger Millet. 9(9): 641–649. |
| [35] | Ueno O, Kawano Y, Wakayama M, and Takeda T. 2006. Leaf vascular systems in C3 and C4 grasses: a two-dimensional analysis Annals of Botany, 97: 611–621. |
| [36] | Ulaganathan and Nirmalakumari, A. 2015. Finger millet germplasm characterization and evaluation using principal component analysis. SABRAO. J. Breed. Genet. 47(2): 79–88. |
| [37] | Vandana B., Rajendra P., and Prabha Sh.. 2021. Genetic variability and correlation studies for morphological and seed quality parameters in foxtail millet. The Pharma Innovation Journal, 10(4), 160–165. |
| [38] | Wossen Tarekegne, Firew Mekbib, and Yigzaw Dessalegn. 2019. Performance and Participatory Variety Evaluation of Finger Millet East African Journal of Sciences. Vol. 13(1) 27-38. |
APA Style
Chimdi, A., Tesso, B., Daba, C. (2024). Genetic Variability Analysis for Yield and Yield-Associated Traits of Finger Millet [Eleusine coracana (L.) Gaertn.] Accessions at Mechara, Eastern Ethiopia. International Journal of Biomedical Science and Engineering, 12(1), 10-18. https://doi.org/10.11648/j.ijbse.20241201.12
ACS Style
Chimdi, A.; Tesso, B.; Daba, C. Genetic Variability Analysis for Yield and Yield-Associated Traits of Finger Millet [Eleusine coracana (L.) Gaertn.] Accessions at Mechara, Eastern Ethiopia. Int. J. Biomed. Sci. Eng. 2024, 12(1), 10-18. doi: 10.11648/j.ijbse.20241201.12
AMA Style
Chimdi A, Tesso B, Daba C. Genetic Variability Analysis for Yield and Yield-Associated Traits of Finger Millet [Eleusine coracana (L.) Gaertn.] Accessions at Mechara, Eastern Ethiopia. Int J Biomed Sci Eng. 2024;12(1):10-18. doi: 10.11648/j.ijbse.20241201.12
@article{10.11648/j.ijbse.20241201.12,
author = {Ababa Chimdi and Bulti Tesso and Chemeda Daba},
title = {Genetic Variability Analysis for Yield and Yield-Associated Traits of Finger Millet [Eleusine coracana (L.) Gaertn.] Accessions at Mechara, Eastern Ethiopia
},
journal = {International Journal of Biomedical Science and Engineering},
volume = {12},
number = {1},
pages = {10-18},
doi = {10.11648/j.ijbse.20241201.12},
url = {https://doi.org/10.11648/j.ijbse.20241201.12},
eprint = {https://article.sciencepublishinggroup.com/pdf/10.11648.j.ijbse.20241201.12},
abstract = {The genetic enhancement of any crop, including finger millet, requires a certain degree of genetic variation for effective utilization in crop improvement programs. However, in Ethiopia, there is limited information on the extent and pattern of genetic variability of finger millet collections under diversified agro-climatic conditions. This makes it difficult for a population to adapt to changing environmental conditions. As a result, the population may be more vulnerable to extinction, exposure to new diseases, low productivity, and selection acting on any genes that may provide disease resistance. Therefore, knowledge of genetic variability is crucial for breeders in order to develop new cultivars with desired traits that are beneficial for both farmers and breeders. The present investigation was carried out to estimating the genetic variability, heritability, and genetic advance in sixty four finger millet accessions for yield and yield-related traits at Mechara agricultural research center. The experimental design was laid out in 8 x 8 simple lattice design. The analysis of variance for mean sum of squares due to genotypes revealed highly significant differences for all the 17 quantitative characters. The genotypes showed the highest mean performance for grain yield ranged from 1.38 ton per hectare for ACC#208448 to 4.35 ton per hectare for ACC#230255. Whereas, genotypic and phenotypic coefficients of variation were found high for biomass yield, harvest index, and grain yield. Broad-sense heritability ranged from 50.12% for the number of fingers per ear to 93.18% for days to heading. High heritability coupled with high genetic advance as percent of mean were observed for leaf number, finger length, ear weight, thousand grain weight, biomass yield, and harvest index. In general, the results demonstrated that the finger millet accessions exhibited a high degree of genetic variability for the traits studied, which can be helpful for genetic enhancement.
},
year = {2024}
}
TY - JOUR T1 - Genetic Variability Analysis for Yield and Yield-Associated Traits of Finger Millet [Eleusine coracana (L.) Gaertn.] Accessions at Mechara, Eastern Ethiopia AU - Ababa Chimdi AU - Bulti Tesso AU - Chemeda Daba Y1 - 2024/06/06 PY - 2024 N1 - https://doi.org/10.11648/j.ijbse.20241201.12 DO - 10.11648/j.ijbse.20241201.12 T2 - International Journal of Biomedical Science and Engineering JF - International Journal of Biomedical Science and Engineering JO - International Journal of Biomedical Science and Engineering SP - 10 EP - 18 PB - Science Publishing Group SN - 2376-7235 UR - https://doi.org/10.11648/j.ijbse.20241201.12 AB - The genetic enhancement of any crop, including finger millet, requires a certain degree of genetic variation for effective utilization in crop improvement programs. However, in Ethiopia, there is limited information on the extent and pattern of genetic variability of finger millet collections under diversified agro-climatic conditions. This makes it difficult for a population to adapt to changing environmental conditions. As a result, the population may be more vulnerable to extinction, exposure to new diseases, low productivity, and selection acting on any genes that may provide disease resistance. Therefore, knowledge of genetic variability is crucial for breeders in order to develop new cultivars with desired traits that are beneficial for both farmers and breeders. The present investigation was carried out to estimating the genetic variability, heritability, and genetic advance in sixty four finger millet accessions for yield and yield-related traits at Mechara agricultural research center. The experimental design was laid out in 8 x 8 simple lattice design. The analysis of variance for mean sum of squares due to genotypes revealed highly significant differences for all the 17 quantitative characters. The genotypes showed the highest mean performance for grain yield ranged from 1.38 ton per hectare for ACC#208448 to 4.35 ton per hectare for ACC#230255. Whereas, genotypic and phenotypic coefficients of variation were found high for biomass yield, harvest index, and grain yield. Broad-sense heritability ranged from 50.12% for the number of fingers per ear to 93.18% for days to heading. High heritability coupled with high genetic advance as percent of mean were observed for leaf number, finger length, ear weight, thousand grain weight, biomass yield, and harvest index. In general, the results demonstrated that the finger millet accessions exhibited a high degree of genetic variability for the traits studied, which can be helpful for genetic enhancement. VL - 12 IS - 1 ER -