1. Introduction
Establishing individual identification is a critical aspect of forensic investigations, applicable in criminal proceedings, medico-legal matters, and cases involving mass disasters
. DNA analysis may disclose a person’s precise identity as well as information about their physical attributes, ethnicity, place of origin and sex
. Globally, approximately 9.2% of deceased individuals remain unidentified, highlighting the importance of addressing this significant humanitarian crisis
. Determining identity factors such as age and sex has become increasingly important in both criminal and civil cases
| [3] | Hamidi O, Afrasiabi M, Namaki M. GADNN: a revolutionary hybrid deep learning neural network for age and sex determination utilizing cone beam computed tomography images of maxillary and frontal sinuses. BMC Medical Research Methodology. 2024; 24(1): 50. https://doi.org/10.1186/s12874-024-02183-9 |
[3]
. Sex determination in humans involves identifying an individual's biological sex, where humans typically develop as either male or female, depending primarily on the arrangement of sex chromosomes inherited from their parents
| [4] | Ornoy A, Weinstein-Fudim L, Ergaz Z. Methods for Prenatal Sex Determination and Their Importance in Understanding and Prevention of Gender-Related Birth Defects. Intech Open, Book. 2019: 1-21. |
[4]
.
In any society, ensuring the security of life and property is crucial. However, despite the best efforts of police and other security organizations, some criminals manage to evade capture due to the challenges posed by large populations in forensic investigations and the continuous rise in diverse criminal activities
| [5] | Kaya DÖ, Koca Y, Kuzubaş TÜ, Kurtaş Ö, Demir İ, Çetin Gr. Sex Determination Using Data Mining Methods Through Measurements of Ascender and Descender Parts of Letters. The Bulletin of Legal Medicine. 2024; 29(1): 19. https://doi.org/10.17986/blm.1690 |
[5]
. Personal identification in forensics is possible with gender determination
. In situations where alternative kinds of identification are not accessible, it acts as a first step in the identification process
| [7] | Arthanari A, Sureshbabu S, Ramalingam K, Prathap L, Ravindran V. Forensic gender prediction by using mandibular morphometric indices: a panoramic radiograph study. Cureus. 2024; 16(3). https://doi.org/10.7759/cureus.56603 |
[7]
. The X and Y sex chromosomes, consisting of tightly bonded DNA and proteins, serve as the genetic blueprints for the development and functioning of all living organisms, including gender-specific physical traits and body structures
| [4] | Ornoy A, Weinstein-Fudim L, Ergaz Z. Methods for Prenatal Sex Determination and Their Importance in Understanding and Prevention of Gender-Related Birth Defects. Intech Open, Book. 2019: 1-21. |
[4]
. Individuals with two X chromosomes typically develop as females, while those with one X and one Y chromosome usually develop as males
| [1] | Meilana ANST, Auerkari EI. DNA Profiling, Bioinformatics and Databases in Forensics: Human Identification Purposes. Journal La Lifesci. 2024; 5(1): 49-63. https://doi.org/10.37899/journallalifesci.v5i1.1171 |
| [4] | Ornoy A, Weinstein-Fudim L, Ergaz Z. Methods for Prenatal Sex Determination and Their Importance in Understanding and Prevention of Gender-Related Birth Defects. Intech Open, Book. 2019: 1-21. |
[1, 4]
.
Including the sex parameter as a supporting element in forensic analysis can enhance the reliability of results and help detectives find accurate matches from a large pool of candidates
| [5] | Kaya DÖ, Koca Y, Kuzubaş TÜ, Kurtaş Ö, Demir İ, Çetin Gr. Sex Determination Using Data Mining Methods Through Measurements of Ascender and Descender Parts of Letters. The Bulletin of Legal Medicine. 2024; 29(1): 19. https://doi.org/10.17986/blm.1690 |
[5]
. Sex assessment methods are typically divided into two main types: morphoscopic (visual or morphological) and metric techniques
| [8] | Curate F. The estimation of sex of human skeletal remains in the Portuguese identified collections: history and prospects. Forensic Sciences. 2022; 2(1): 272-86. https://www.mdpi.com/2673-6756/2/1/21# |
[8]
. Morphoscopic methods tend to be more subjective because they rely on the observer's interpretation, making them prone to bias (
| [8] | Curate F. The estimation of sex of human skeletal remains in the Portuguese identified collections: history and prospects. Forensic Sciences. 2022; 2(1): 272-86. https://www.mdpi.com/2673-6756/2/1/21# |
| [9] | Tekeli E, Gültekin T, Doksanaltı ME, Öztaner SH, Elma C. Accurate sex determination using ancient DNA analysis for human skeletal remains from different historical archeological sites in Turkey. Mediterranean Archaeology and Archaeometry, Vol. 20, No 1, (2020), pp. 93-106. https://doi.org/10.5281/zenodo.3605672 |
[8, 9]
). Traditional morphological and morphometric analyses can determine sex, but these methods are unreliable for infant and juvenile or fragmentary adult skeletons
| [6] | Prasad P, Jaber M, Ramani P, Arafat A, Khairy A. SRY gene isolation from teeth for forensic gender identification—An observational study. Plos one. 2024; 19(1): e0294751. https://doi.org/10.1371/journal.pone.0297835 |
| [8] | Curate F. The estimation of sex of human skeletal remains in the Portuguese identified collections: history and prospects. Forensic Sciences. 2022; 2(1): 272-86. https://www.mdpi.com/2673-6756/2/1/21# |
| [9] | Tekeli E, Gültekin T, Doksanaltı ME, Öztaner SH, Elma C. Accurate sex determination using ancient DNA analysis for human skeletal remains from different historical archeological sites in Turkey. Mediterranean Archaeology and Archaeometry, Vol. 20, No 1, (2020), pp. 93-106. https://doi.org/10.5281/zenodo.3605672 |
| [10] | Sume BW. Estimation of body height from percutaneous length of tibia in Debre Markos University students, North West Ethiopia. Egyptian Journal of Forensic Sciences. 2019; 9: 1-8. https://doi.org/10.1186/s41935-019-0157-z |
[6, 8-10]
.
Significantly, biochemical methods developed in the late 20th century possess the highest accuracy and reliability in sex estimation
| [11] | Heng D, Manica S, Franco A. Forensic Dentistry as an Analysis Tool for Sex Estimation: A Review of Current Techniques. Research and Reports in Forensic Medical Science. 2022: 25-39. https://doi.org/10.2147/RRFMS.S334796 |
[11]
. Forensic DNA typing and subsequent molecular methods of sex determination in humans have proven to be essential tools in the criminal justice system
| [12] | Dash HR, Rawat N, Das S. Alternatives to amelogenin markers for sex determination in humans and their forensic relevance. Molecular biology reports. 2020; 47(3): 2347-60. https://doi.org/10.1007/s11033-020-05268-y |
[12]
. DNA analysis for sex determination is particularly useful because it can be applied to samples lacking morphological characteristics
| [13] | Albernaz Neves J, Antunes-Ferreira N, Machado V, Botelho J, Proença L, Quintas A, et al. An umbrella review of the evidence of sex determination procedures in forensic dentistry. Journal of Personalized Medicine. 2022; 12(5): 787. https://doi.org/10.3390/jpm12050787 |
[13]
. Genetic sex determination methods are not dependent on subjective physical examination, are highly accurate, require only small sample sizes, and do not necessitate the evaluation of specific tissues, allowing any organ to be used
| [14] | AL-Ameedy QS, Al-Khafaji NSK. Discrimination the Gender in the Criminal Evidence at Crime Scene. Indian Journal of Forensic Medicine & Toxicology. 2020; 14(2): 1853-7. |
[14]
.
Advances in technology and molecular biology, particularly the Polymerase Chain Reaction (PCR), offer the most sensitive, accurate, and rapid technique for sex determination by analyzing gender-specific sequences on the X and Y chromosomes
| [15] | Salih MH. Molecular markers for human sex determination in forensic genetics analysis. International Journal for Research in Applied Sciences and Biotechnology. 2021; 8(6): 25-30. https://doi.org/10.31033/ijrasb.8.6.6 |
[15]
. The presence of a Y chromosome in males, which is detected by specific sequences, allows for the distinction between male and female individuals
. Genetic markers for sex determination based on PCR analysis include various loci, notably the SRY (sex-determining region Y) and amelogenin (AMEL)
| [15] | Salih MH. Molecular markers for human sex determination in forensic genetics analysis. International Journal for Research in Applied Sciences and Biotechnology. 2021; 8(6): 25-30. https://doi.org/10.31033/ijrasb.8.6.6 |
[15]
.
The AMEL gene, also known as the enamel protein gene, is located in the Xp22.1-Xp22.3, Yp11.2 region of the X and Y chromosome respectively
. It is ideal for genetic sexing because it amplifies a short intron 1 region, generating approximately 100 bp products that differ in length between the X and Y chromosomes
. It was first described in 1993
| [19] | Gabriele A, Chierto E, Gino S, Inturri S, Aneli S, Robino C. Privacy and ethical challenges of the Amelogenin sex test in forensic paternity/kinship analysis: Insights from a 13-year case history. Forensic Science International: Synergy. 2023; 7: 100440. https://doi.org/10.1016/j.fsisyn.2023.100440 |
[19]
. AMEL, a major matrix protein found in human enamel, exhibits different signatures (or sizes and patterns of the nucleotide sequence) in males and females
. The AMEL gene encoding female AMEL is located on the X chromosome (AMEL X), while the AMEL gene encoding male AMEL is located on the Y chromosome (AMEL Y)
. A 6 bp deletion in intron 1 of the X chromosome results in a shorter amplicon compared to the Y chromosome. Consequently, female DNA (XX) produces identical-length products, while male DNA (XY) generates products differing by 6 bp
.
Every object at the crime scene has an important meaning as forensic evidence
| [21] | Kurniawan A, Rizky BN, Prakoeswa BF, Athalia SA, Malau ST, Alias A. The significance of amelogenin loci from toothpicks as forensic evidence for sex determination. Journal of Taibah University Medical Sciences. 2023; 18(1): 148-53. https://doi.org/10.1016/j.jtumed.2022.07.010 |
[21]
. Blood samples are the ideal form for biological evidence that are collected from different kinds of crimes i.e. murder, rape-sexual homicide, hit and run, road accident cases, tool marks, or other types of heinous crime
| [15] | Salih MH. Molecular markers for human sex determination in forensic genetics analysis. International Journal for Research in Applied Sciences and Biotechnology. 2021; 8(6): 25-30. https://doi.org/10.31033/ijrasb.8.6.6 |
[15]
. However, teeth and facial bones display remarkable resilience against various destructive forces such as fire, burns, and even highly concentrated acids, making them viable for recovery from disaster sites
| [22] | Lim JJ-Y, Khamis, M. F., Abd Rashid, N. H.. Identification of amelogenin gene on burnt teeth samples through nested polymerase chain reaction amplification for sex identification. Sains Kesihatan Malaysia (2019) 17(01): 91-8. http://dx.doi.org./10.17576/JSKM-2019-1701-12 |
| [23] | Lim JJ-Y, Khamis MF, Abd Rashid NHB. Application of Two Sex Markers by Nested PCR for Gender Determination. Indian Journal of Forensic Medicine & Toxicology. 2021; 15(2): 2636-42. https://doi.org/10.37506/ijfmt.v15i2.14769 |
[22, 23]
. With the progress in PCR techniques, extracting DNA from dental pulp and other tooth structures has become more straightforward. Consequently, accurate genetic identification of individuals has become more feasible
. AMEL test is also suitable for determining the sex of highly degraded DNA because it generates short products
. Therefore, Genetic methods are reliable, do not require living cells, and can obtain DNA from very ancient and nonviable tissues, making these methods the most widely accepted
| [14] | AL-Ameedy QS, Al-Khafaji NSK. Discrimination the Gender in the Criminal Evidence at Crime Scene. Indian Journal of Forensic Medicine & Toxicology. 2020; 14(2): 1853-7. |
[14]
.
Sex typing can be performed using primers that specifically amplify the region of the AMEL locus
. Common AMEL primers produce amplicons of 106 and 112 bp or 212 and 218 bp for the X and Y loci, respectively, which are then separated by electrophoresis
| [25] | Kristanto R. Genetic Analysis of Genealogy from Dental Y-STR DNA with Raman Spectra Method: Literature Review. Indian Journal of Forensic Medicine & Toxicology. 2022; 16(4): 395-401. |
[25]
. However, several limitations of the AMEL sex test have emerged, in particular, the efficacy of the AMEL marker subsequently came into question when cases of false identification of males as females started surfacing
| [11] | Heng D, Manica S, Franco A. Forensic Dentistry as an Analysis Tool for Sex Estimation: A Review of Current Techniques. Research and Reports in Forensic Medical Science. 2022: 25-39. https://doi.org/10.2147/RRFMS.S334796 |
| [19] | Gabriele A, Chierto E, Gino S, Inturri S, Aneli S, Robino C. Privacy and ethical challenges of the Amelogenin sex test in forensic paternity/kinship analysis: Insights from a 13-year case history. Forensic Science International: Synergy. 2023; 7: 100440. https://doi.org/10.1016/j.fsisyn.2023.100440 |
[11, 19]
. It was hypothesized that this could be due to mutation in the primer-binding region, or deletion of the AMEL Y gene in males that led to possibly erroneous conclusions
| [11] | Heng D, Manica S, Franco A. Forensic Dentistry as an Analysis Tool for Sex Estimation: A Review of Current Techniques. Research and Reports in Forensic Medical Science. 2022: 25-39. https://doi.org/10.2147/RRFMS.S334796 |
[11]
.
Nevertheless, the AMEL test is included in practically all commercial human identification kits, which allows genetic sexing in addition to determining identity
. Therefore, this review aimed to comprehensively evaluate the progress made in utilizing AMEL gene analysis for sex determination, including advancements in methodology and techniques, while also addressing the existing challenges hindering its widespread adoption and effectiveness.
2. Methodology
2.1. Search Strategy
A thorough search was conducted to evaluate progress in utilizing AMEL gene analysis for sex determination, focusing on advancements in methodology and techniques, while addressing existing challenges. Databases used for searching were Cochrane Library, PubMed, and Google Scholar with specified key phrases and Boolean operators (AND, OR, NOT). Main keywords included "amelogenin gene," "sex determination," "gender identification," "sexual dimorphism," "sex marker," "advancements," "challenges," "forensic," "forensic genetics," "molecular analysis," and "genetic analysis." This strategy aimed to identify relevant articles that assess both the advancements and challenges of AMEL gene-based sex determination in forensic contexts.
2.2. Eligibility Criteria
A comprehensive systematic review adhering to the PRISMA (Preferred Reporting Items for Systematic Reviews and Meta-Analysis) guidelines was conducted to explore advancements and challenges in AMEL gene-based sex determination. The review employed a focused research question framed within the PICo framework, aiming to evaluate the advancements and challenges of using AMEL gene analysis for sex determination. Specifically, it examined the advancements and challenges of AMEL gene analysis (Interest) in determining sex in human samples (Population/Problem/Condition) within forensic science applications (Context/Setting). The review aimed to provide a thorough assessment of both the advancements and challenges of this method in forensic contexts
| [26] | Zissler A, Stoiber W, Steinbacher P, Geissenberger J, Monticelli FC, Pittner S. Postmortem protein degradation as a tool to estimate the PMI: A systematic review. Diagnostics. 2020; 10(12): 1014. https://doi.org/10.3390/diagnostics10121014 |
[26]
.
Table 1. PICo (population, interest, context/setting) format.
Component | Description |
Population | Individuals or samples undergoing sex determination through amelogenin gene analysis |
Interest | Effectiveness and challenges of using amelogenin gene analysis for sex determination |
Context | Forensic science setting/ context |
2.2.1. Inclusion and Exclusion Criteria
Stringent inclusion and exclusion criteria were applied to ensure the selection of relevant and high-quality research articles. Only research articles for forensic application, focusing explicitly on human subjects, published between 2019 and 2024, in English, and with full text availability were considered. The studies needed to address sex determination using the AMEL gene as a biomarker.
Conversely, non-human studies and articles not available in full text or published in languages other than English were excluded. Additionally, studies focusing on sex markers other than the AMEL gene were not considered. This systematic approach ensured that the review incorporated only studies directly pertinent to the topic, maintaining consistency and accessibility throughout the chosen literature.
2.2.2. Study Selection
The study selection process meticulously followed established criteria to ensure relevance and quality. Initially, a comprehensive database search was conducted, identifying and eliminating duplicate records. The remaining articles were systematically evaluated by scrutinizing titles and abstracts, focusing on study relevance to forensic sciences and sex determination through AMEL gene analysis. Articles not directly related to forensic sciences or lacking relevance to AMEL gene-based sex determination were excluded. Research articles pertinent to the subject were retained for further assessment. Full-text versions of these retained studies were then carefully examined for final eligibility.
Only studies explicitly addressing sex determination through AMEL gene analysis were included, while those focusing on different biomarkers or unrelated topics were excluded. This stringent selection process ensured that the systematic review comprised relevant and high-quality studies, contributing significantly to the understanding of AMEL gene-based sex determination in forensic contexts.
2.3. Data Extraction
The authors were carefully extracted data from each chosen article to ensure accuracy and completeness. A detailed dataset was created, including important information like the first author's name, publication year, sample details, DNA concentration, treatment methods, extraction techniques, analytical approaches, result, and conclusions. Any unclear or missing information was noted to maintain transparency. This thorough data extraction process allowed for a comprehensive analysis of the articles and accurate capture of key insights. The following table provides a summary of the articles included in the study on sex determination using AMEL gene analysis.
2.4. Quality Assessment
The quality and potential biases of a study are critical factors in assessing the significance and reliability of its outcomes. To evaluate the internal validity of the selected articles, the authors developed specific signaling questions based on the Cochrane Risk of Bias tool (RoB 2.0)
. Drawing on a framework established by Zissler et al. (2020), a model was created to estimate the overall quality and risk of bias in the included studies
| [26] | Zissler A, Stoiber W, Steinbacher P, Geissenberger J, Monticelli FC, Pittner S. Postmortem protein degradation as a tool to estimate the PMI: A systematic review. Diagnostics. 2020; 10(12): 1014. https://doi.org/10.3390/diagnostics10121014 |
[26]
. These signaling questions focused on five key domains: appropriate sample sizes, the inclusion of challenging forensic samples, treatment of samples under varying conditions, the presence of confirmatory tests, and the robustness of analytical techniques. The risk of bias and methodological quality for each study were independently assessed by the review authors, with items rated as having low, moderate, or high risk of bias. Studies that employed rigorous controls, utilized well-established extraction and analytical techniques, and transparently reported their methods were rated more highly. Conversely, studies with unclear methodologies or incomplete data were critically evaluated, with careful consideration given to their impact on the overall conclusions of the review (Detail in Appendix I and II)
2.5. Data Analysis
Data analysis involved comparing and contrasting the findings from the various studies included in the review. This analysis focused on identifying trends and patterns in the effectiveness of different DNA extraction methods and analytical techniques used for AMEL gene analysis. Particular attention was given to evaluating the sensitivity and specificity of these methods across different sample types and conditions. The analysis also explored the success rates of sex determination using AMEL gene analysis, particularly in challenging forensic contexts where samples might be degraded or present in low concentrations.
Furthermore, the analysis considered the reliability of AMEL gene analysis when used alone versus in combination with other markers, such as SRY or Y-STR. Studies were grouped based on the type of sample and extraction method used, and their results were compared to assess the overall effectiveness of these approaches. The findings were then synthesized to provide insights into the best practices for AMEL gene analysis in forensic and biological research, highlighting areas where further research or methodological improvements might be needed.
Figure 1. Flow chart of the literature search process and study selection according to PRISMA guidelines.
3. Result
3.1. Study Selection
The systematic search on PubMed and Cochrane databases retrieved 1,030 records, with an additional 12 studies found through Google Scholar using various keyword combinations. After removing 336 duplicates, 706 studies were screened by titles and abstracts. From these, 664 articles were excluded for not meeting the inclusion criteria, mainly due to their focus on non-forensic content or non-human research. Of the remaining 42 studies, 34 were further excluded for not focusing on sex determination or using different markers like SRY, STR, SNPs (single nucleotide polymorphism), or hormonal assays. Reference examinations led to 2 additional relevant study, resulting in a total of 10 studies included in the review. A PRISMA diagram illustrates the article selection process
| [28] | Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. Bmj. 2021; 372. https://doi.org/10.1136/bmj.n71 |
[28]
. To generate Boolean operators for searches in PubMed and Cochrane, the following phrases were combined with “AND”. (Amelogenin or sex identification) AND (forensic OR challenges OR advancement), (amelogenin or sex determination) AND (forensic OR challenges OR advancement), (amelogenin or sex marker) AND (forensic OR challenges for advancement) (
Figure 1).
3.2. Data Extraction
The authors have carefully extracted data from each chosen article to ensure accuracy and completeness. A detailed dataset was created, including important information like the first author's name, publication year, sample details, DNA concentration, treatment methods, extraction techniques, analytical approaches, result, and conclusions. Any unclear or missing information was noted to maintain transparency. This thorough data extraction process allowed for a comprehensive analysis of the articles and accurate capture of key insights. The following table provides a summary of the articles included in the study on sex determination using AMEL gene analysis (
Table 2).
Table 2. Studies summary on sex determination using AMEL analysis.
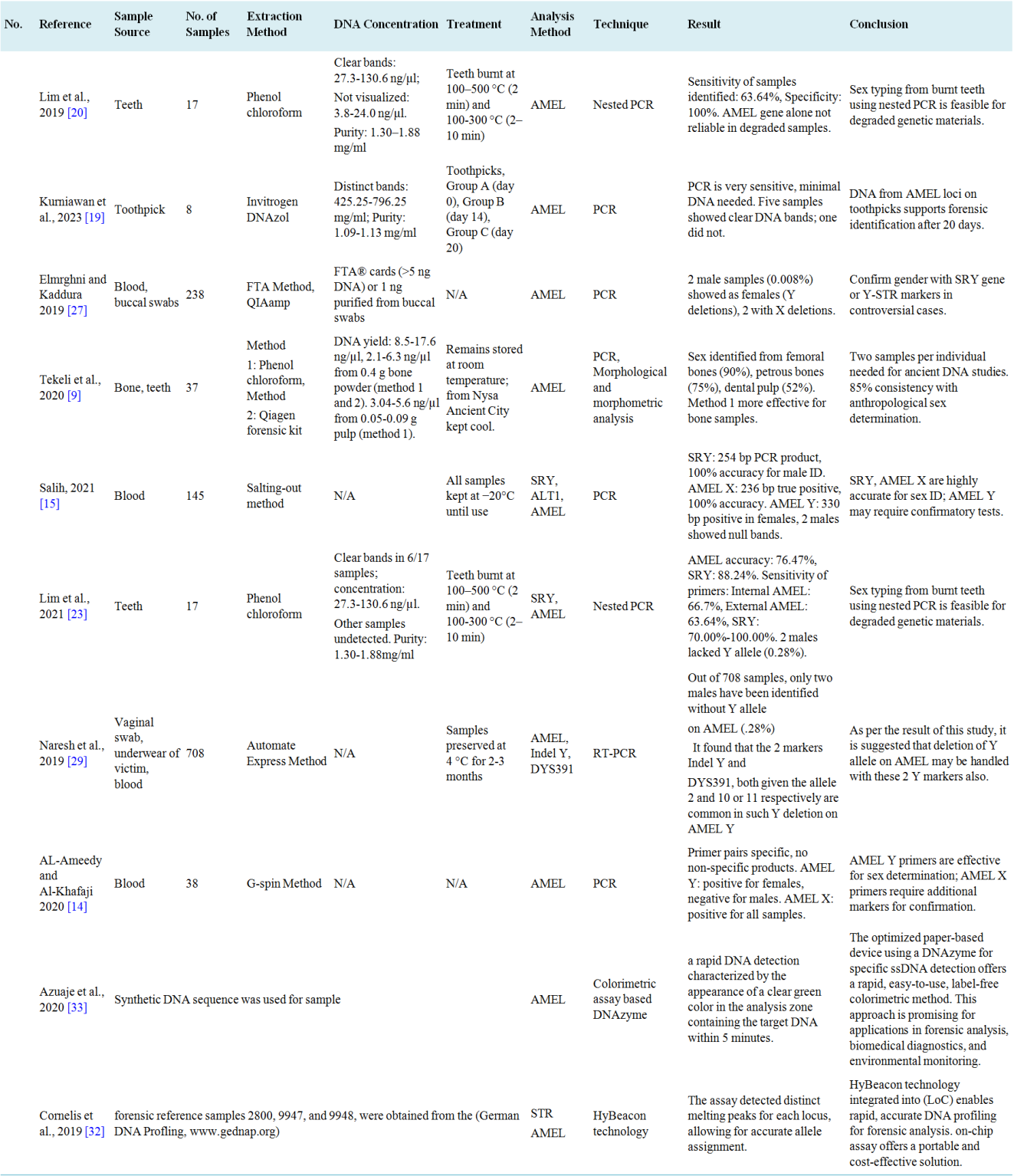
3.3. Study Characteristics
The ten studies included in this review exhibit considerable variation in several key parameters. The studies were conducted in different geographical regions, including Germany, Spain, Turkey, Malaysia, India, Indonesia, Iraq, and Libya, providing a global perspective on the advancements and challenges of AMEL gene-based sex determination in forensic contexts. The studies covered a range of sample sizes, from small case studies to larger population-based analyses, and the biological samples analyzed include blood (two studies
| [14] | AL-Ameedy QS, Al-Khafaji NSK. Discrimination the Gender in the Criminal Evidence at Crime Scene. Indian Journal of Forensic Medicine & Toxicology. 2020; 14(2): 1853-7. |
| [15] | Salih MH. Molecular markers for human sex determination in forensic genetics analysis. International Journal for Research in Applied Sciences and Biotechnology. 2021; 8(6): 25-30. https://doi.org/10.31033/ijrasb.8.6.6 |
[14, 15]
), teeth (two studies
| [22] | Lim JJ-Y, Khamis, M. F., Abd Rashid, N. H.. Identification of amelogenin gene on burnt teeth samples through nested polymerase chain reaction amplification for sex identification. Sains Kesihatan Malaysia (2019) 17(01): 91-8. http://dx.doi.org./10.17576/JSKM-2019-1701-12 |
| [23] | Lim JJ-Y, Khamis MF, Abd Rashid NHB. Application of Two Sex Markers by Nested PCR for Gender Determination. Indian Journal of Forensic Medicine & Toxicology. 2021; 15(2): 2636-42. https://doi.org/10.37506/ijfmt.v15i2.14769 |
[22, 23]
), vaginal swabs and victim's underwear blood (one study
| [29] | Naresh K, Chauhan A, Gupta R. Role of 3-Y markers in Y deleted allele on amelogenin in autosomal STR. Int J Mol Biol Open Access. 2019; 4(2): 75-8. |
[29]
), a toothpick (one study
| [21] | Kurniawan A, Rizky BN, Prakoeswa BF, Athalia SA, Malau ST, Alias A. The significance of amelogenin loci from toothpicks as forensic evidence for sex determination. Journal of Taibah University Medical Sciences. 2023; 18(1): 148-53. https://doi.org/10.1016/j.jtumed.2022.07.010 |
[21]
), both blood and buccal swabs (one study
), and both bone and teeth (one study
| [9] | Tekeli E, Gültekin T, Doksanaltı ME, Öztaner SH, Elma C. Accurate sex determination using ancient DNA analysis for human skeletal remains from different historical archeological sites in Turkey. Mediterranean Archaeology and Archaeometry, Vol. 20, No 1, (2020), pp. 93-106. https://doi.org/10.5281/zenodo.3605672 |
[9]
). It was observed that, the reviewed studies mainly utilized samples from blood and teeth, with others including vaginal swabs, victim's underwear, buccal swabs, and toothpicks. This may be because, blood samples are ideal for biological evidence from various crimes, such as murder, rape, hit-and-run, road accidents, and other heinous offenses
| [15] | Salih MH. Molecular markers for human sex determination in forensic genetics analysis. International Journal for Research in Applied Sciences and Biotechnology. 2021; 8(6): 25-30. https://doi.org/10.31033/ijrasb.8.6.6 |
[15]
. Teeth are highly suitable for DNA analysis in sex identification, especially from fragmented, decomposed, and burnt corpses due to their high mineralization and resistance to heat and decomposition
| [22] | Lim JJ-Y, Khamis, M. F., Abd Rashid, N. H.. Identification of amelogenin gene on burnt teeth samples through nested polymerase chain reaction amplification for sex identification. Sains Kesihatan Malaysia (2019) 17(01): 91-8. http://dx.doi.org./10.17576/JSKM-2019-1701-12 |
[22]
.
Isolation of genomic DNA is the key step for numerous applications and molecular studies ranging from basic research to routine diagnostic and therapeutic decision-making. A wide variety of techniques have been employed to extract DNA from different sources
| [31] | Suresh1 S, KB, SB. DNA Extraction from Archived Paraffin Embedded Tissues: A Comparative Study Using Three Different Extraction Method-Extraction of DNA from Paraffin Embedded Tissues. Medico-legal Update, October-December 2020; Vol. 20, No. 4. |
[31]
. Among the included studies, two studies used the phenol-chloroform extraction method
| [22] | Lim JJ-Y, Khamis, M. F., Abd Rashid, N. H.. Identification of amelogenin gene on burnt teeth samples through nested polymerase chain reaction amplification for sex identification. Sains Kesihatan Malaysia (2019) 17(01): 91-8. http://dx.doi.org./10.17576/JSKM-2019-1701-12 |
| [23] | Lim JJ-Y, Khamis MF, Abd Rashid NHB. Application of Two Sex Markers by Nested PCR for Gender Determination. Indian Journal of Forensic Medicine & Toxicology. 2021; 15(2): 2636-42. https://doi.org/10.37506/ijfmt.v15i2.14769 |
[22, 23]
, one employed the salting-out method
| [15] | Salih MH. Molecular markers for human sex determination in forensic genetics analysis. International Journal for Research in Applied Sciences and Biotechnology. 2021; 8(6): 25-30. https://doi.org/10.31033/ijrasb.8.6.6 |
[15]
, another utilized the DNAZol method
| [21] | Kurniawan A, Rizky BN, Prakoeswa BF, Athalia SA, Malau ST, Alias A. The significance of amelogenin loci from toothpicks as forensic evidence for sex determination. Journal of Taibah University Medical Sciences. 2023; 18(1): 148-53. https://doi.org/10.1016/j.jtumed.2022.07.010 |
[21]
, one combined FTA with QIAamp
, one paired phenol-chloroform with the Qiagen Forensic Kit
| [9] | Tekeli E, Gültekin T, Doksanaltı ME, Öztaner SH, Elma C. Accurate sex determination using ancient DNA analysis for human skeletal remains from different historical archeological sites in Turkey. Mediterranean Archaeology and Archaeometry, Vol. 20, No 1, (2020), pp. 93-106. https://doi.org/10.5281/zenodo.3605672 |
[9]
, another used the G-Spin method
| [14] | AL-Ameedy QS, Al-Khafaji NSK. Discrimination the Gender in the Criminal Evidence at Crime Scene. Indian Journal of Forensic Medicine & Toxicology. 2020; 14(2): 1853-7. |
[14]
, and one relied on the Automate Express method
| [29] | Naresh K, Chauhan A, Gupta R. Role of 3-Y markers in Y deleted allele on amelogenin in autosomal STR. Int J Mol Biol Open Access. 2019; 4(2): 75-8. |
[29]
. Regarding AMEL gene analysis, four studies used standard PCR
| [14] | AL-Ameedy QS, Al-Khafaji NSK. Discrimination the Gender in the Criminal Evidence at Crime Scene. Indian Journal of Forensic Medicine & Toxicology. 2020; 14(2): 1853-7. |
| [15] | Salih MH. Molecular markers for human sex determination in forensic genetics analysis. International Journal for Research in Applied Sciences and Biotechnology. 2021; 8(6): 25-30. https://doi.org/10.31033/ijrasb.8.6.6 |
| [21] | Kurniawan A, Rizky BN, Prakoeswa BF, Athalia SA, Malau ST, Alias A. The significance of amelogenin loci from toothpicks as forensic evidence for sex determination. Journal of Taibah University Medical Sciences. 2023; 18(1): 148-53. https://doi.org/10.1016/j.jtumed.2022.07.010 |
| [30] | Elmrghni S, Kaddura M. Human identification by amelogenin test in Libyans. Am J Biomed Sci & Res AJBSR. 2019; 3(6). http://dx.doi.org/10.34297/AJBSR.2019.03.000737 |
[14, 15, 21, 30]
, two employed nested PCR
| [22] | Lim JJ-Y, Khamis, M. F., Abd Rashid, N. H.. Identification of amelogenin gene on burnt teeth samples through nested polymerase chain reaction amplification for sex identification. Sains Kesihatan Malaysia (2019) 17(01): 91-8. http://dx.doi.org./10.17576/JSKM-2019-1701-12 |
| [23] | Lim JJ-Y, Khamis MF, Abd Rashid NHB. Application of Two Sex Markers by Nested PCR for Gender Determination. Indian Journal of Forensic Medicine & Toxicology. 2021; 15(2): 2636-42. https://doi.org/10.37506/ijfmt.v15i2.14769 |
[22, 23]
, one utilized Real time PCR (RT-PCR)
| [29] | Naresh K, Chauhan A, Gupta R. Role of 3-Y markers in Y deleted allele on amelogenin in autosomal STR. Int J Mol Biol Open Access. 2019; 4(2): 75-8. |
[29]
, another combined PCR with morphological and morphometric techniques
| [9] | Tekeli E, Gültekin T, Doksanaltı ME, Öztaner SH, Elma C. Accurate sex determination using ancient DNA analysis for human skeletal remains from different historical archeological sites in Turkey. Mediterranean Archaeology and Archaeometry, Vol. 20, No 1, (2020), pp. 93-106. https://doi.org/10.5281/zenodo.3605672 |
[9]
), one study used integration of PCR amplification and HyBeacon melting assays
| [32] | Cornelis S, Tytgat O, Fauvart M, Gansemans Y, Vander Plaetsen A-S, Wiederkehr RS, et al. Silicon µPCR chip for forensic STR profiling with hybeacon probe melting curves. Scientific Reports. 2019; 9(1): 7341. https://doi.org/10.1038/s41598-019-43946-5 |
[32]
, and the other calorimetric assay using DNAzymes in paper devices
| [33] | Azuaje-Hualde E, Arroyo-Jimenez S, Garai-Ibabe G, de Pancorbo MM, Benito-Lopez F, Basabe-Desmonts L. Naked eye Y amelogenin gene fragment detection using DNAzymes on a paper-based device. Analytica Chimica Acta. 2020; 1123: 1-8. https://doi.org/10.1016/j.aca.2020.05.010 |
[33]
. Key findings from these studies will be discussed in the subsequent sections of this review.
3.4. Quality Of Included Articles
The quality assessment of the included articles reveals a diverse range of risk of bias, largely influenced by sample size, the use of challenging forensic samples, and the application of confirmatory tests. Studies like Naresh et al., (2019) demonstrated a low risk of bias due to their large sample size, use of challenging forensic samples (e.g., vaginal swabs and underwear), and robust confirmatory tests
| [29] | Naresh K, Chauhan A, Gupta R. Role of 3-Y markers in Y deleted allele on amelogenin in autosomal STR. Int J Mol Biol Open Access. 2019; 4(2): 75-8. |
[29]
. However, many studies, including Lim et al., (2019), Tekeli et al., (2020), and Salih (2021), fell into the moderate risk of bias category
| [9] | Tekeli E, Gültekin T, Doksanaltı ME, Öztaner SH, Elma C. Accurate sex determination using ancient DNA analysis for human skeletal remains from different historical archeological sites in Turkey. Mediterranean Archaeology and Archaeometry, Vol. 20, No 1, (2020), pp. 93-106. https://doi.org/10.5281/zenodo.3605672 |
[9]
. These studies often included adequate sample sizes and employed standard analytical techniques but lacked consistent use of confirmatory tests, which is crucial in forensic analysis.
On the other hand, studies such as Kurniawan et al., (2023) and Azuaje et al., (2020) exhibited a high risk of bias due to small or undefined sample sizes, the absence of challenging forensic samples, and insufficient confirmatory tests
| [21] | Kurniawan A, Rizky BN, Prakoeswa BF, Athalia SA, Malau ST, Alias A. The significance of amelogenin loci from toothpicks as forensic evidence for sex determination. Journal of Taibah University Medical Sciences. 2023; 18(1): 148-53. https://doi.org/10.1016/j.jtumed.2022.07.010 |
| [33] | Azuaje-Hualde E, Arroyo-Jimenez S, Garai-Ibabe G, de Pancorbo MM, Benito-Lopez F, Basabe-Desmonts L. Naked eye Y amelogenin gene fragment detection using DNAzymes on a paper-based device. Analytica Chimica Acta. 2020; 1123: 1-8. https://doi.org/10.1016/j.aca.2020.05.010 |
[21, 33]
). The variability in conditions across studies, including thermal and storage treatments, further influenced the overall quality ratings. Robust analytical techniques like PCR were generally employed across studies, but the lack of confirmatory testing in many cases weakens the reliability of the results, underscoring the need for more stringent validation processes in future research (See supplementary 2 for detailed information).
4. Discussion
Reliable sex determination requires an accuracy rate of 80%, with most DNA analysis methods achieving up to 100% accuracy
. AMEL is a sex typing marker that is commonly utilized
| [21] | Kurniawan A, Rizky BN, Prakoeswa BF, Athalia SA, Malau ST, Alias A. The significance of amelogenin loci from toothpicks as forensic evidence for sex determination. Journal of Taibah University Medical Sciences. 2023; 18(1): 148-53. https://doi.org/10.1016/j.jtumed.2022.07.010 |
| [35] | Siddique N, Shahid AA, Sughra K. Diversification of Pakistani Amelogenin-Y-Null Male Haplotypes. Scientifica. 2021; 2021(1): 5521411. https://doi.org/10.1155/2021/5521411 |
[21, 35]
. It is present on both X and Y chromosomes where it is denoted as AMEL-X and AMEL-Y. AMEL-X is located on the distal short arm of the X chromosome in the p22.1–p22.3 region, while AMEL-Y is present near the centromere of the Y chromosome at p11.2
. Using primers specific for parts of intron 1 of the gene, fragments of the gene sequence can be amplified
. The two most commonly used sets of AMEL primers gives 106 and 112 bp or 212 and 218 bp amplification product (amplicon) for the AMEL X and AMEL Y loci, respectively. The distinguishing feature is a 6-base pair deletion in intron 1 of the AMEL gene on the X chromosome, allowing differentiation between PCR-amplified products of the gene on the X and Y chromosomes
| [20] | Liu S, Zeng Y, Wang C, Zhang Q, Chen M, Wang X, et al. seGMM: A new tool for gender determination from massively parallel sequencing data. Frontiers in Genetics. 2022; 13: 850804. https://doi.org/10.3389/fgene.2022.850804 |
[20]
.
4.1. DNA Extraction
The effectiveness of AMEL gene analysis is significantly influenced by the source of the sample, as different sample types yield varying amounts and qualities of DNA, which in turn impacts the reliability of the results. Teeth samples, as highlighted by Lim et al., (2021), provide a variable yield of DNA when extracted using the Phenol Chloroform method, with concentrations ranging from 27.3 to 130.6 ng/μl
| [23] | Lim JJ-Y, Khamis MF, Abd Rashid NHB. Application of Two Sex Markers by Nested PCR for Gender Determination. Indian Journal of Forensic Medicine & Toxicology. 2021; 15(2): 2636-42. https://doi.org/10.37506/ijfmt.v15i2.14769 |
[23]
. This variability suggests that while teeth can be a reliable source of DNA, the quality and quantity of DNA obtained can fluctuate depending on the extraction method and the condition of the teeth. Similarly, Mishra et al., (2020) found that organic extraction methods produced complete genetic profiles in 70% of teeth samples, indicating that teeth are a viable source for AMEL gene analysis, though results may vary depending on the extraction protocol used
| [36] | Mishra IK, Mishra A, Kharkwal AC, Mohapatra BK, Behera C, Singh B. Comparative Study on Different Modified Techniques Used For DNA Isolation From Teeth Samples for Obtaining Optimized Output. Indian Journal of Forensic Medicine & Toxicology. 2020; 14(3). |
[36]
.
In comparison, bone samples tend to produce lower DNA yields than teeth, especially when using different extraction methods. Tekeli et al., (2020) reported DNA yields of 8.5-17.6 ng/µl from bone powder using the Phenol Chloroform method and even lower yields of 2.1-6.3 ng/µl with the Qiagen forensic kit. Despite the lower yields, bone samples, particularly femoral bones, have shown high success rates (90%) in sex identification, making them a reliable source for AMEL gene analysis when higher yields are not critical
| [9] | Tekeli E, Gültekin T, Doksanaltı ME, Öztaner SH, Elma C. Accurate sex determination using ancient DNA analysis for human skeletal remains from different historical archeological sites in Turkey. Mediterranean Archaeology and Archaeometry, Vol. 20, No 1, (2020), pp. 93-106. https://doi.org/10.5281/zenodo.3605672 |
[9]
. However, the lower DNA yields from bones highlight the importance of selecting an efficient extraction method to ensure sufficient DNA for analysis.
Buccal swabs and FTA cards represent less invasive sampling methods but typically yield lower DNA concentrations compared to teeth and bone samples. Elmrghni and Kaddura (2019) found that FTA cards provided DNA yields of more than 5 ng, while buccal swabs yielded around 1 ng, which is significantly lower
. This lower yield makes buccal swabs and FTA cards less ideal for AMEL gene analysis, especially in cases requiring high sensitivity or where the DNA sample might be degraded.
Formalin-fixed paraffin-embedded (FFPE) tissues, such as those from archival oral squamous cell carcinoma samples, present another challenging source for DNA extraction. A study comparing different extraction methods found that the salting-out method yielded significantly higher DNA quantities than both the commercial kit and microwave methods
| [31] | Suresh1 S, KB, SB. DNA Extraction from Archived Paraffin Embedded Tissues: A Comparative Study Using Three Different Extraction Method-Extraction of DNA from Paraffin Embedded Tissues. Medico-legal Update, October-December 2020; Vol. 20, No. 4. |
[31]
. Nevertheless, the DNA from FFPE tissues often faces degradation challenges, making it less reliable for AMEL gene analysis compared to fresher or less processed samples like teeth or bones. Similarly, Study by…viewed as FFPE is less ideal for Next Generation Sequencing (NGS) compared to fresh-frozen (FF) samples, which is because the nucleic acids extracted from FFPE tissues tend to be fragmented, limited in quantity, and may contain formalin-induced modifications and mutations
| [37] | Mathieson W, Thomas G. Using FFPE tissue in genomic analyses: advantages, disadvantages and the role of biospecimen science. Current Pathobiology Reports. 2019; 7: 35-40. https://doi.org/10.1007/s40139-019-00194-6 |
[37]
. However, new gender identification markers were added to subsequent STR kits. One widely used kit, the GlobalFiler (GF) PCR amplification kit by Applied Biosystems, which yield higher quality DNA
| [38] | Takayama T. A novel mutation at the AMEL primer binding region on the Y chromosome in AMELY negative male. International Journal of Legal Medicine. 2022; 136(2): 519-26. https://doi.org/10.1007/s00414-022-02781-6 |
[38].
In summary, teeth and bone samples generally provide more reliable DNA for AMEL gene analysis, with teeth offering higher and more variable yields depending on the extraction method. Bone samples, while yielding less DNA, can still provide high accuracy in sex determination. In contrast, buccal swabs, FTA cards, and FFPE tissues tend to yield lower DNA amounts, making them less suitable for high-sensitivity applications unless coupled with highly efficient extraction methods. Selecting the appropriate sample type and extraction method is crucial for optimizing the yield and quality of DNA necessary for accurate AMEL gene analysis.
4.2. Techniques for Analyzing the Amelogenin Gene
Various studies have employed different types of PCR to analyze the AMEL gene for sex determination in forensic and biological samples, each highlighting the strengths and limitations of these methods. Kurniawan et al., (2023) demonstrated the high sensitivity of conventional PCR, which requires minimal DNA to produce clear DNA bands in five out of six samples. This finding underscores the utility of conventional PCR in detecting DNA from minimal and potentially compromised samples, such as toothpicks, even after 20 days, showcasing its robustness in forensic identification
| [21] | Kurniawan A, Rizky BN, Prakoeswa BF, Athalia SA, Malau ST, Alias A. The significance of amelogenin loci from toothpicks as forensic evidence for sex determination. Journal of Taibah University Medical Sciences. 2023; 18(1): 148-53. https://doi.org/10.1016/j.jtumed.2022.07.010 |
[21]
. In contrast, Elmrghni and Kaddura (2019) also used conventional PCR but encountered issues with Y deletions, leading to the misidentification of two male samples as female. Additionally, X deletions were noted in two other samples, suggesting that conventional PCR of the AMEL gene alone might not be sufficient for accurate sex determination in cases where genetic anomalies are present. To mitigate these issues, the study recommends using the SRY gene or Y-STR markers for confirmation in controversial cases, highlighting the limitations of relying solely on AMEL gene analysis
.
Tekeli et al., (2020) took a different approach by comparing conventional PCR with morphological and morphometric analysis, focusing on different bone samples. Their findings indicated varying success rates in sex identification, with femoral bones yielding a 90% success rate, petrous bones 75%, and dental pulp 52%. The study concluded that using multiple samples per individual is essential for ancient DNA studies and emphasized the importance of integrating multiple methods to achieve more reliable results
| [9] | Tekeli E, Gültekin T, Doksanaltı ME, Öztaner SH, Elma C. Accurate sex determination using ancient DNA analysis for human skeletal remains from different historical archeological sites in Turkey. Mediterranean Archaeology and Archaeometry, Vol. 20, No 1, (2020), pp. 93-106. https://doi.org/10.5281/zenodo.3605672 |
[9]
. Meanwhile, Salih (2021) compared conventional PCR analysis of the SRY and AMEL genes, finding that the SRY gene had a 100% accuracy rate for male identification, while the AMEL X gene was equally accurate. However, the AMEL Y gene produced false positives in females and null bands in two males, pointing to potential issues with relying on AMEL Y alone. This study suggests that while SRY and AMEL X are highly reliable, AMEL Y results should be confirmed with additional tests for accurate sex determination
| [15] | Salih MH. Molecular markers for human sex determination in forensic genetics analysis. International Journal for Research in Applied Sciences and Biotechnology. 2021; 8(6): 25-30. https://doi.org/10.31033/ijrasb.8.6.6 |
[15]
.
In contrast to conventional PCR, Naresh et al., (2019) utilized real-time PCR (RT-PCR) for AMEL gene analysis
| [29] | Naresh K, Chauhan A, Gupta R. Role of 3-Y markers in Y deleted allele on amelogenin in autosomal STR. Int J Mol Biol Open Access. 2019; 4(2): 75-8. |
[29]
. RT-PCR offers the advantage of rapid detection, allowing amplified DNA to be detected in less than an hour through the combination of PCR chemistry with fluorescent probe detection
. Their study found that, two male samples out of 708 (0.28%) lacking the Y allele on the AMEL gene. To address such cases, they utilized Indel Y and DYS391 markers, which are commonly associated with AMEL Y allele deletions
| [29] | Naresh K, Chauhan A, Gupta R. Role of 3-Y markers in Y deleted allele on amelogenin in autosomal STR. Int J Mol Biol Open Access. 2019; 4(2): 75-8. |
[29]
. These findings highlight the importance of incorporating multiple Y-specific markers for accurate and reliable sex determination in rare instances of AMEL Y allele deletions. Similarly, AL-Ameedy and Al-Khafaji (2020) demonstrated the specificity of primer pairs in conventional PCR, with no non-specific products observed. Their results showed that AMEL Y primers were effective for sex determination, being positive in females and negative in males, while AMEL X primers were positive for all samples. However, they concluded that AMEL X primers require additional markers for confirmation, indicating the need for a comprehensive approach to achieve accurate results
| [14] | AL-Ameedy QS, Al-Khafaji NSK. Discrimination the Gender in the Criminal Evidence at Crime Scene. Indian Journal of Forensic Medicine & Toxicology. 2020; 14(2): 1853-7. |
[14]
.
Lim et al., (2019) explored the application of nested PCR, a more refined technique involving two rounds of amplification with different primers, where the product of the first round serves as the template for the second round
. This method increases the sensitivity of PCR by allowing a higher number of cycles, which is particularly beneficial for analyzing degraded genetic materials. Their study, which focused on sex typing from burnt teeth, found that nested PCR was feasible for degraded genetic materials, identifying 76.47% of samples with a sensitivity of 63.64% and specificity of 100%. However, they also noted that the AMEL gene alone is not reliable in degraded samples, necessitating additional confirmatory tests
| [22] | Lim JJ-Y, Khamis, M. F., Abd Rashid, N. H.. Identification of amelogenin gene on burnt teeth samples through nested polymerase chain reaction amplification for sex identification. Sains Kesihatan Malaysia (2019) 17(01): 91-8. http://dx.doi.org./10.17576/JSKM-2019-1701-12 |
[22]
). Lim et al., (2021) reinforced these findings, reporting similar sensitivity and specificity rates with nested PCR, where six out of seventeen samples showed clear bands and DNA concentrations ranging from 27.3 to 130.6 ng/µl
| [23] | Lim JJ-Y, Khamis MF, Abd Rashid NHB. Application of Two Sex Markers by Nested PCR for Gender Determination. Indian Journal of Forensic Medicine & Toxicology. 2021; 15(2): 2636-42. https://doi.org/10.37506/ijfmt.v15i2.14769 |
[23]
. The study further supports the use of nested PCR for sex typing in degraded samples. Kristanto (2022) added that nested PCR enhances the sensitivity of AMEL gene analysis by 9.5-fold, based on the AMEL ratio before and after nested PCR, demonstrating the effectiveness of this method for analyzing degraded DNA
| [25] | Kristanto R. Genetic Analysis of Genealogy from Dental Y-STR DNA with Raman Spectra Method: Literature Review. Indian Journal of Forensic Medicine & Toxicology. 2022; 16(4): 395-401. |
[25]
.
A recent advancement in on-site DNA fingerprinting features the AMEL gender determination assay, moving towards portable forensic tools
| [32] | Cornelis S, Tytgat O, Fauvart M, Gansemans Y, Vander Plaetsen A-S, Wiederkehr RS, et al. Silicon µPCR chip for forensic STR profiling with hybeacon probe melting curves. Scientific Reports. 2019; 9(1): 7341. https://doi.org/10.1038/s41598-019-43946-5 |
[32]
. The optimized in situ DNA detection method in a paper device uses a DNAzyme targeting the Y human AMEL fragment for rapid, non-enzymatic, colorimetric detection, quantitatively analysis by calculating grey intensity values, while qualitative detection is indicated by a green "coffee ring. DNA sensing probes and hemin bind to the Y fragment, forming a catalytic structure that oxidizes ABTS (2,2'-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid) with hydrogen peroxide(H
2O
2), producing a green color on the paper support
| [33] | Azuaje-Hualde E, Arroyo-Jimenez S, Garai-Ibabe G, de Pancorbo MM, Benito-Lopez F, Basabe-Desmonts L. Naked eye Y amelogenin gene fragment detection using DNAzymes on a paper-based device. Analytica Chimica Acta. 2020; 1123: 1-8. https://doi.org/10.1016/j.aca.2020.05.010 |
[33]
. Likewise, according to (Cornelis et al., 2019), a probe fully complementary to the Y allele with a 2-nucleotide mismatch to the X allele enables differentiation of genotypes by measuring the multing temprature(Tm) of the HyBeacon probe-amelogenin duplex. Controlled heating of PCR product from 40 °C to 65 °C shows a 10 °C lower Tm for the X allele due to the mismatch. This difference in Tm facilitates unbiased gender identification
| [32] | Cornelis S, Tytgat O, Fauvart M, Gansemans Y, Vander Plaetsen A-S, Wiederkehr RS, et al. Silicon µPCR chip for forensic STR profiling with hybeacon probe melting curves. Scientific Reports. 2019; 9(1): 7341. https://doi.org/10.1038/s41598-019-43946-5 |
[32]
.
In summary, while conventional PCR is robust and widely applicable, especially for minimal and compromised samples, its accuracy can be compromised by genetic anomalies, necessitating supplementary markers. Real-time PCR offers rapid detection but requires careful consideration of primer sensitivity. Nested PCR, with its higher sensitivity, is particularly effective for degraded samples, making it a valuable tool in forensic DNA analysis, although it too may require confirmatory tests for comprehensive results. However, the integration of PCR amplification and HyBeacon melting assays, and the use of DNAzymes in paper devices have highlighted advancements in gender identification.
4.3. Challenges in Sex Determination Using Amelogenin Gene
The efficacy of the AMEL marker has been questioned due to cases where males were falsely identified as females. This issue is hypothesized to stem from mutations in the primer-binding region or deletions of the AMEL Y gene in males, leading to erroneous conclusions
| [11] | Heng D, Manica S, Franco A. Forensic Dentistry as an Analysis Tool for Sex Estimation: A Review of Current Techniques. Research and Reports in Forensic Medical Science. 2022: 25-39. https://doi.org/10.2147/RRFMS.S334796 |
[11]
. The analysis is further complicated by forensic samples often containing degraded DNA, which can hinder or even prevent accurate sex determination
| [15] | Salih MH. Molecular markers for human sex determination in forensic genetics analysis. International Journal for Research in Applied Sciences and Biotechnology. 2021; 8(6): 25-30. https://doi.org/10.31033/ijrasb.8.6.6 |
| [34] | Maulani C, Auerkari EI. Molecular analysis for sex determination in forensic dentistry: a systematic review. Egyptian Journal of Forensic Sciences. 2020; 10: 1-9. https://doi.org/10.1186/s41935-020-00210-6 |
[15, 34]
. (Lim et al., 2019) noted that while the method is highly specific, with a specificity of 100%, its sensitivity of 63.64% needs improvement
| [22] | Lim JJ-Y, Khamis, M. F., Abd Rashid, N. H.. Identification of amelogenin gene on burnt teeth samples through nested polymerase chain reaction amplification for sex identification. Sains Kesihatan Malaysia (2019) 17(01): 91-8. http://dx.doi.org./10.17576/JSKM-2019-1701-12 |
[22]
. Challenges with samples showing no DNA bands, were encountered, indicating potential issues with DNA degradation or extraction inefficiency
| [21] | Kurniawan A, Rizky BN, Prakoeswa BF, Athalia SA, Malau ST, Alias A. The significance of amelogenin loci from toothpicks as forensic evidence for sex determination. Journal of Taibah University Medical Sciences. 2023; 18(1): 148-53. https://doi.org/10.1016/j.jtumed.2022.07.010 |
| [22] | Lim JJ-Y, Khamis, M. F., Abd Rashid, N. H.. Identification of amelogenin gene on burnt teeth samples through nested polymerase chain reaction amplification for sex identification. Sains Kesihatan Malaysia (2019) 17(01): 91-8. http://dx.doi.org./10.17576/JSKM-2019-1701-12 |
| [23] | Lim JJ-Y, Khamis MF, Abd Rashid NHB. Application of Two Sex Markers by Nested PCR for Gender Determination. Indian Journal of Forensic Medicine & Toxicology. 2021; 15(2): 2636-42. https://doi.org/10.37506/ijfmt.v15i2.14769 |
[21-23]
. The use of unconventional sample sources like toothpicks also presents challenges due to their limited DNA yield
| [21] | Kurniawan A, Rizky BN, Prakoeswa BF, Athalia SA, Malau ST, Alias A. The significance of amelogenin loci from toothpicks as forensic evidence for sex determination. Journal of Taibah University Medical Sciences. 2023; 18(1): 148-53. https://doi.org/10.1016/j.jtumed.2022.07.010 |
[21]
.
Additionally, (Lim et al., 2021) noted the relatively low sensitivity of primers for the internal and external AMEL loci (6.67% and 63.64%, respectively), highlighting the difficulties in amplifying this gene in highly degraded samples
| [23] | Lim JJ-Y, Khamis MF, Abd Rashid NHB. Application of Two Sex Markers by Nested PCR for Gender Determination. Indian Journal of Forensic Medicine & Toxicology. 2021; 15(2): 2636-42. https://doi.org/10.37506/ijfmt.v15i2.14769 |
[23]
. Further, AL-Ameedy and Al-Khafaji (2020) reported that while AMEL Y primers effectively differentiate between male and female samples, AMEL X primers showed positive results for all samples, indicating a need for careful interpretation
| [14] | AL-Ameedy QS, Al-Khafaji NSK. Discrimination the Gender in the Criminal Evidence at Crime Scene. Indian Journal of Forensic Medicine & Toxicology. 2020; 14(2): 1853-7. |
[14]
). However, according to (Lim et al., 2021) nested PCR has shown promise in overcoming some of these challenges. It was reported that only 7 out of 17 samples were successfully amplified with the AMEL external primer due to degraded DNA, but subsequent amplification with the AMEL internal primer succeeded in 13 out of 17 samples
| [23] | Lim JJ-Y, Khamis MF, Abd Rashid NHB. Application of Two Sex Markers by Nested PCR for Gender Determination. Indian Journal of Forensic Medicine & Toxicology. 2021; 15(2): 2636-42. https://doi.org/10.37506/ijfmt.v15i2.14769 |
[23]
. This indicates that nested PCR, by amplifying smaller DNA fragments, can improve the chances of successful sex typing in degraded samples.
Again, Elmrghni and Kaddura (2019) highlighted the limitations of the AMEL test, where deletions in the Y chromosome led to false female identifications in 0.008% of cases (2 out of 238 males)
. Similarly, Naresh et al., (2019) also highlighted that, out of 708 male samples, 0.28% lacked the Y allele on AMEL gene
| [29] | Naresh K, Chauhan A, Gupta R. Role of 3-Y markers in Y deleted allele on amelogenin in autosomal STR. Int J Mol Biol Open Access. 2019; 4(2): 75-8. |
[29]
. However, new gender identification markers were added to subsequent STR kits. One widely used kit, the GlobalFiler (GF) PCR amplification kit by Applied Biosystems, includes the AMEL marker along with two additional markers, Y-indel and DYS391. These additions have significantly lowered the risk of misidentifying AMELY-negative males as female
| [38] | Takayama T. A novel mutation at the AMEL primer binding region on the Y chromosome in AMELY negative male. International Journal of Legal Medicine. 2022; 136(2): 519-26. https://doi.org/10.1007/s00414-022-02781-6 |
[38]
. This emphasizes the necessity of using multiple sex markers such as Indel Y, SRY, and DYS391 to ensure accurate sex determination
| [23] | Lim JJ-Y, Khamis MF, Abd Rashid NHB. Application of Two Sex Markers by Nested PCR for Gender Determination. Indian Journal of Forensic Medicine & Toxicology. 2021; 15(2): 2636-42. https://doi.org/10.37506/ijfmt.v15i2.14769 |
| [29] | Naresh K, Chauhan A, Gupta R. Role of 3-Y markers in Y deleted allele on amelogenin in autosomal STR. Int J Mol Biol Open Access. 2019; 4(2): 75-8. |
[23, 29]
. Further, Tekeli et al., (2020) discussed the variability in DNA yield from ancient remains, finding higher yields from femoral bones compared to petrous bones and dental pulp
| [9] | Tekeli E, Gültekin T, Doksanaltı ME, Öztaner SH, Elma C. Accurate sex determination using ancient DNA analysis for human skeletal remains from different historical archeological sites in Turkey. Mediterranean Archaeology and Archaeometry, Vol. 20, No 1, (2020), pp. 93-106. https://doi.org/10.5281/zenodo.3605672 |
[9]
.
Lastly, the optimized paper device uses a DNAzyme targeting the Y AMEL fragment for rapid, non-enzymatic, colorimetric DNA detection, with limits of 655 ng in solution and 45.7 ng on paper
| [33] | Azuaje-Hualde E, Arroyo-Jimenez S, Garai-Ibabe G, de Pancorbo MM, Benito-Lopez F, Basabe-Desmonts L. Naked eye Y amelogenin gene fragment detection using DNAzymes on a paper-based device. Analytica Chimica Acta. 2020; 1123: 1-8. https://doi.org/10.1016/j.aca.2020.05.010 |
[33]
. Despite the improvement, the 45.7 ng limit is still high for detecting very low concentrations, indicating a need for further optimization. Further modifications or alternative methods are needed to lower the detection limit. Generally, these studies collectively underscore the inherent limitations of relying solely on the AMEL gene for sex determination and highlight the need for complementary markers, methodologies, and techniques to enhance accuracy and reliability in forensic cases.
Appendix
Appendix I: Assessment of Risk of Bias
The review authors developed a tailored tool to evaluate the potential risk of bias in the studies included in this review. This tool, adapted from the Cochrane Risk of Bias (RoB) 2.0 tool, focuses on five key domains relevant to the studies analyzed. Each domain addresses critical aspects that could influence the validity and reliability of the study outcomes. The tool is designed to guide reviewers in assessing the methodological quality and potential biases within these studies.
Key Domains of Bias:
Appropriate Sample Sizes
Inclusion of Challenging Forensic Samples
Treatment of Samples Under Varying Conditions
Presence of Confirmatory Tests
Robustness of Analytical Techniques
For each domain, specific signaling questions were developed to facilitate judgments about the risk of bias. The responses to these signaling questions are used to determine the overall quality of each study, which is categorized as high risk of bias, moderate risk of bias, or low risk of bias.
Response Options:
Y: Yes
PY: Probably Yes
N: No
PN: Probably No
NI: No Information
The "probably" responses indicate that a judgment has been made by the reviewer, while "no information" is used only when insufficient data are available to make a reasonable judgment. Each response contributes to a "domain-level judgment" and an "overall quality" rating for each study.
Judgment Options:
Low Risk of Bias
Moderate Risk of Bias
High Risk of Bias
Signaling Questions:
1. Appropriate Sample Sizes
1.1 Was the sample size sufficient to appropriately address the research hypothesis? (Y/PY/N/PN/NI)
A sufficient sample size is critical for ensuring that the study’s findings are representative of the population being studied. A “Yes” indicates that the sample size was adequate for the intended analysis.
2. Inclusion of Challenging Forensic Samples
2.1 Were challenging forensic samples included in the study? (Y/PY/N/PN/NI)
Including challenging samples, such as those that are degraded or have been exposed to adverse conditions, is essential for assessing the robustness of the study's methods. A “Yes” indicates that such samples were appropriately included.
3. Treatment of Samples Under Varying Conditions
3.1 Were samples treated under varying conditions to assess the robustness of the methods? (Y/PY/N/PN/NI)
This question evaluates whether the study considered different treatment conditions (e.g., temperature, time) to ensure that the methods are reliable under various scenarios. A “Yes” indicates comprehensive treatment of samples.
4. Presence of Confirmatory Tests
4.1 Were confirmatory tests conducted to validate the findings? (Y/PY/N/PN/NI)
Confirmatory tests are crucial for verifying the results and ensuring the reliability of the study's conclusions. A “Yes” indicates that such tests were performed and reported.
5. Robustness of Analytical Techniques
5.1 Were the analytical techniques robust and well-established? (Y/PY/N/PN/NI)
Robust analytical techniques are necessary to produce reliable and reproducible results. This question assesses whether the study employed established methods that are appropriate for the analysis. A “Yes” indicates that robust techniques were used.
Appendix II: Quality Assessment of Included Studies
Evaluation of risk of bias assessment for each of the studies included in the review based on the provided key domains and signaling questions
1. Lim et al., 2019
Sample Size (1.1): Y - Sample size of 17 is reasonable for preliminary analysis.
Challenging Forensic Samples (2.1): Y - Used burnt teeth, which represent challenging samples.
Varying Conditions (3.1): Y - Samples treated under varying temperatures and time.
Confirmatory Tests (4.1): N - No confirmatory tests mentioned.
Robust Analytical Techniques (5.1): PY - Nested PCR is established but may not be ideal for degraded samples.
Overall Quality Rating: Moderate Risk of Bias
2. Kurniawan et al., 2023
Sample Size (1.1): N - Sample size of 8 is small for generalizability.
Challenging Forensic Samples (2.1): N - Toothpicks may not represent a true challenging sample.
Varying Conditions (3.1): Y - Samples were treated under varying storage conditions.
Confirmatory Tests (4.1): NI - No information on confirmatory tests provided.
Robust Analytical Techniques (5.1): Y - PCR is a well-established method.
Overall Quality Rating: High Risk of Bias
3. Elmrghni and Kaddura 2019
Sample Size (1.1): Y - A larger sample size (238) is sufficient for analysis.
Challenging Forensic Samples (2.1): Y - Blood and buccal swabs can be challenging in certain contexts.
Varying Conditions (3.1): NI - No information provided on varying treatment conditions.
Confirmatory Tests (4.1): Y - Suggestion of confirmatory tests for controversial cases.
Robust Analytical Techniques (5.1): Y - Established FTA and QIAamp methods were used.
Overall Quality Rating: Moderate Risk of Bias
4. Tekeli et al., 2020
Sample Size (1.1): Y - Sample size (37) appears adequate for the analysis.
Challenging Forensic Samples (2.1): Y - Used bone and teeth, which are challenging.
Varying Conditions (3.1): Y - Samples were kept under different conditions.
Confirmatory Tests (4.1): N - No confirmatory tests mentioned.
Robust Analytical Techniques (5.1): Y - Techniques used were well-established.
Overall Quality Rating: Moderate Risk of Bias
5. Salih, 2021
Sample Size (1.1): Y - Adequate sample size (145).
Challenging Forensic Samples (2.1): N - Blood samples alone may not be particularly challenging.
Varying Conditions (3.1): Y - Samples were stored under controlled conditions.
Confirmatory Tests (4.1): Y - Confirmatory tests were outlined for ambiguous results.
Robust Analytical Techniques (5.1): Y - Standard methods were employed.
Overall Quality Rating: Moderate Risk of Bias
6. Lim et al., 2021
Sample Size (1.1): Y - Sample size of 17 is adequate for exploratory analysis.
Challenging Forensic Samples (2.1): Y - Similar to Lim et al., 2019, used burnt teeth.
Varying Conditions (3.1): Y - Varied thermal conditions used for testing.
Confirmatory Tests (4.1): N - No mention of confirmatory tests.
Robust Analytical Techniques (5.1): PY - Nested PCR, reviewed previously.
Overall Quality Rating: Moderate Risk of Bias
7. Naresh et al., 2019
Sample Size (1.1): Y - Adequate sample size (708).
Challenging Forensic Samples (2.1): Y - Includes vaginal swabs and underwear, which can be challenging.
Varying Conditions (3.1): Y - Samples preserved at specific conditions.
Confirmatory Tests (4.1): Y - Reliability of AMEL results benefits from additional confirmatory tests.
Robust Analytical Techniques (5.1): Y - RT-PCR is a robust method.
Overall Quality Rating: Low Risk of Bias
8. AL-Ameedy and Al-Khafaji 2020
Sample Size (1.1): N - Small sample size (38), limits generalizability.
Challenging Forensic Samples (2.1): N - Blood does not represent significant challenge.
Varying Conditions (3.1): NI - No information on varying treatment conditions.
Confirmatory Tests (4.1): Y - Confirms need for additional markers for reliability.
Robust Analytical Techniques (5.1): Y - Established methods used.
Overall Quality Rating: Moderate Risk of Bias
9. Azuaje et al., 2020
Sample Size (1.1): NI - No sample size mentioned; unclear if adequate.
Challenging Forensic Samples (2.1): NI - Synthetic DNA may not apply to real forensic situations.
Varying Conditions (3.1): NI - Not applicable as no actual samples or conditions described.
Confirmatory Tests (4.1): NI - No confirmatory tests addressed.
Robust Analytical Techniques (5.1): Y - Promising technique discussed.
Overall Quality Rating: High Risk of Bias
10. Cornelis et al., 2019
Sample Size (1.1): Y - Sufficient sample sizes (2800 reference samples).
Challenging Forensic Samples (2.1): N - Reference samples may not be considered challenging.
Varying Conditions (3.1): NI - No information provided on varied conditions.
Confirmatory Tests (4.1): Y - Accurate allele assignment suggests confirmatory processes.
Robust Analytical Techniques (5.1): Y - HyBeacon is known for effective application in forensics.
Overall Quality Rating: Moderate Risk of Bias
Table 3. Summary of Risk of Bias Assessments.
Reference | 1.1 | 2.1 | 3.1 | 4.1 | 5.1 | Overall Risk of Bias |
Lim et al., 2019 | PY | Y | Y | N | PY | Moderate |
Kurniawan et al., 2023 | PN | Y | Y | N | PN | High |
Elmrghni and Kaddura, 2019 | Y | PY | NI | Y | PN | Moderate |
Tekeli et al., 2020 | PY | Y | Y | PN | PN | Moderate |
Salih, 2021 | Y | PN | PY | Y | PN | Moderate |
Lim et al., 2021 | PY | Y | Y | Y | PY | Moderate |
Naresh et al., 2019 | Y | Y | Y | Y | PY | Low |
AL-Ameedy and Al-Khafaji, 2020 | PY | PN | NI | N | PN | Moderate |
Azuaje et al., 2020 | NI | N | NI | PN | Y | High |
Cornelis et al., 2019 | Y | N | NI | Y | Y | Moderate |
